Correlation Heatmap with Hierarchical Clustering in Python
- 10 Aug 2025
- Acadmic Method, Machine Learning
- by alasbahimoha
Summary
This article walks you through building a “top-tier-journal–ready” correlation heatmap with hierarchical clustering in Python to analyze complex relationships between two groups of variables (e.g., microbes vs. environmental factors). You’ll learn to use Seaborn and SciPy to:
- Automatically compute Spearman correlations and p-values
- Add significance stars (
***for p < 0.01,**for 0.01–0.05) - Cluster both rows and columns to reveal hidden structure
- Save a high-resolution figure ready for publication
We’ll follow a clear workflow: read two worksheets from an Excel file (microbial data & environmental factors), align samples (rows) to keep only shared samples, compute pairwise Spearman r and p, sanitize any NaNs, render a clustered heatmap with significance markers, and export the figure.
Code (step-by-step) 💻
1) Imports — the foundation
# Core data & math
import pandas as pd
import numpy as np
# Plotting
import seaborn as sns
import matplotlib
import matplotlib.pyplot as plt
# Stats
from scipy.stats import spearmanr
# (Optional GUI backend if you run locally outside notebooks)
# matplotlib.use('TkAgg')
Why these libraries?
- pandas / numpy: efficient tables & numeric ops
- seaborn / matplotlib: beautiful, customizable plots; we’ll use
seaborn.clustermap - scipy.stats.spearmanr: Spearman’s rank correlation and p-values
2) Data alignment & preparation
We must ensure row alignment (sample IDs) between the two tables before computing correlations.
def plot_spearman_clustermap(data1: pd.DataFrame, data2: pd.DataFrame,
out_path: str = "clustered_correlation.png",
vmax: float = 0.5):
"""
Build a clustered heatmap of Spearman correlations between
variables in data1 (rows) and variables in data2 (cols),
with significance stars from p-values.
Parameters
----------
data1 : DataFrame
Samples x variables (e.g., microbes)
data2 : DataFrame
Samples x variables (e.g., environmental factors)
out_path : str
Where to save the figure
vmax : float
Color scale half-range; heatmap uses [-vmax, +vmax]
"""
# --- 1) Align on row index (sample IDs) ---
print("\n--- Aligning sample rows between the two datasets ---")
print(f"Before align, data1 first 5 index: {data1.index[:5].tolist()} ...")
print(f"Before align, data2 first 5 index: {data2.index[:5].tolist()} ...")
data1, data2 = data1.align(data2, join='inner', axis=0)
print(f"After align, shared samples: {data1.shape[0]}")
if data1.shape[0] == 0:
raise ValueError(
"Error: The two datasets have no shared sample indices. "
"Check that sample IDs match across sheets."
)
# Names for heatmap axes
rows = data1.columns # will be heatmap rows
cols = data2.columns # will be heatmap cols
# Initialize containers for r and p
corr_matrix = pd.DataFrame(index=rows, columns=cols, dtype=float)
p_matrix = pd.DataFrame(index=rows, columns=cols, dtype=float)
What this does
align(..., join='inner', axis=0)keeps only shared samples (row indices) so every pairwise correlation uses the same samples.- Safety check stops early if there are no shared samples (common source of errors).
Tip: For the alignment to work, both DataFrames must have sample IDs as their index. See the “Main script” section below for recommended reading options.
3) Correlation & significance
We compute Spearman r and p-values for every pair (each column in data1 vs. each column in data2). We also sanitize any residual NaNs.
# --- 2) Compute Spearman r and p for each pair ---
for r in rows:
for c in cols:
corr, p_val = spearmanr(data1[r], data2[c], nan_policy='omit')
corr_matrix.loc[r, c] = corr
p_matrix.loc[r, c] = p_val
# Final cleanup: if any correlations are NaN (e.g., <2 valid points), set to 0
corr_matrix.fillna(0, inplace=True)
Why Spearman?
Spearman uses ranks, making it robust to outliers and nonlinearity (monotonic but not necessarily linear relationships).
About NaNs
Even with nan_policy='omit', if too few valid data points remain, spearmanr can return NaN. We convert those to 0 so plotting won’t crash.
4) Figure — clustermap with stars
We convert the p-value matrix into a star annotation matrix and feed both into seaborn.clustermap. We also tune figure size, fonts, dendrogram lines, and a custom legend for p-value stars.
# --- 3) Build significance star annotations ---
annot_matrix = p_matrix.map(lambda p: '***' if p < 0.01 else ('**' if p < 0.05 else ''))
# Global font tweaks
plt.rcParams['font.sans-serif'] = ['Times New Roman']
plt.rcParams['axes.unicode_minus'] = False
# Dynamic size based on matrix shape
n_rows, n_cols = corr_matrix.shape
fig_width = n_cols * 0.5 + 4
fig_height = n_rows * 0.5 + 4
cmap = 'PuOr_r' # divergent palette (purple–orange, reversed)
# --- 4) Draw clustered heatmap ---
g = sns.clustermap(
corr_matrix,
method='average', # UPGMA
metric='euclidean',
cmap=cmap,
vmin=-vmax, vmax=vmax, # symmetric color scale
annot=annot_matrix,
fmt='s', # annotations are strings (stars)
annot_kws={"size": 16, "color": "black", "fontweight": "bold"},
linewidths=.5, linecolor='white',
dendrogram_ratio=0.1,
figsize=(fig_width, fig_height),
cbar_pos=(1.15, 0.65, 0.03, 0.2),
cbar_kws={'label': "Spearman's r"},
)
# Ticks & labels
plt.setp(g.ax_heatmap.get_xticklabels(), rotation=90, fontsize=12)
plt.setp(g.ax_heatmap.get_yticklabels(), rotation=0, fontsize=12)
# Colorbar label & ticks
g.cax.set_ylabel("Spearman's r", fontsize=12, fontweight='bold')
g.cax.tick_params(labelsize=10)
# Make dendrogram lines slightly thicker for clarity
if g.ax_row_dendrogram.collections:
g.ax_row_dendrogram.collections[0].set_linewidth(1.5)
if g.ax_col_dendrogram.collections:
g.ax_col_dendrogram.collections[0].set_linewidth(1.5)
# --- 5) Manual legend for P-value stars ---
legend_text = "P value\n*** < 0.01\n** 0.01–0.05"
plt.text(1.15, 0.6, legend_text, transform=g.fig.transFigure,
fontsize=11, va='top',
bbox=dict(boxstyle='round,pad=0.5', fc='white', ec='black', lw=1))
# Save & show
plt.savefig(out_path, bbox_inches='tight', dpi=300, pad_inches=0.2)
plt.show()
return g
What you get
- A clustered heatmap of Spearman r values
- Row/column dendrograms from average linkage clustering
- Significance stars overlaid on each cell
- A clean colorbar and a compact legend for p-values
5) Main script — the “control panel”
This is the only part you need to edit to run on your own data. It reads two sheets, sets the correct indices (sample IDs), and calls the plotting function.
if __name__ == "__main__":
# >>> EDIT THESE THREE LINES FOR YOUR DATA <<<
input_excel_path = r"your\full\path\your_data.xlsx"
sheet_name_1 = "Microbial_Guilds" # microbes (rows = samples, cols = taxa/guilds)
sheet_name_2 = "Environmental_Factors" # environment (rows = samples, cols = factors)
print(f"Loading data from '{input_excel_path}' ...")
# Recommended: set the **same** sample ID column as index for BOTH sheets.
# Example: if the first column in each sheet is 'SampleID', do:
microbe_data = pd.read_excel(input_excel_path, sheet_name=sheet_name_1, index_col=0)
env_data = pd.read_excel(input_excel_path, sheet_name=sheet_name_2, index_col=0)
print("Data loaded. Shapes:", microbe_data.shape, env_data.shape)
# Run the plotter (change output path if you like)
plot_spearman_clustermap(microbe_data, env_data,
out_path="clustered_correlation.png",
vmax=0.5)
Why set index_col=0 for both?
It ensures the first column (sample IDs) becomes the row index for both tables, which makes align(..., axis=0) work exactly as intended. If your IDs are in a different column, pass that column index or name to index_col.
How to use with your own data
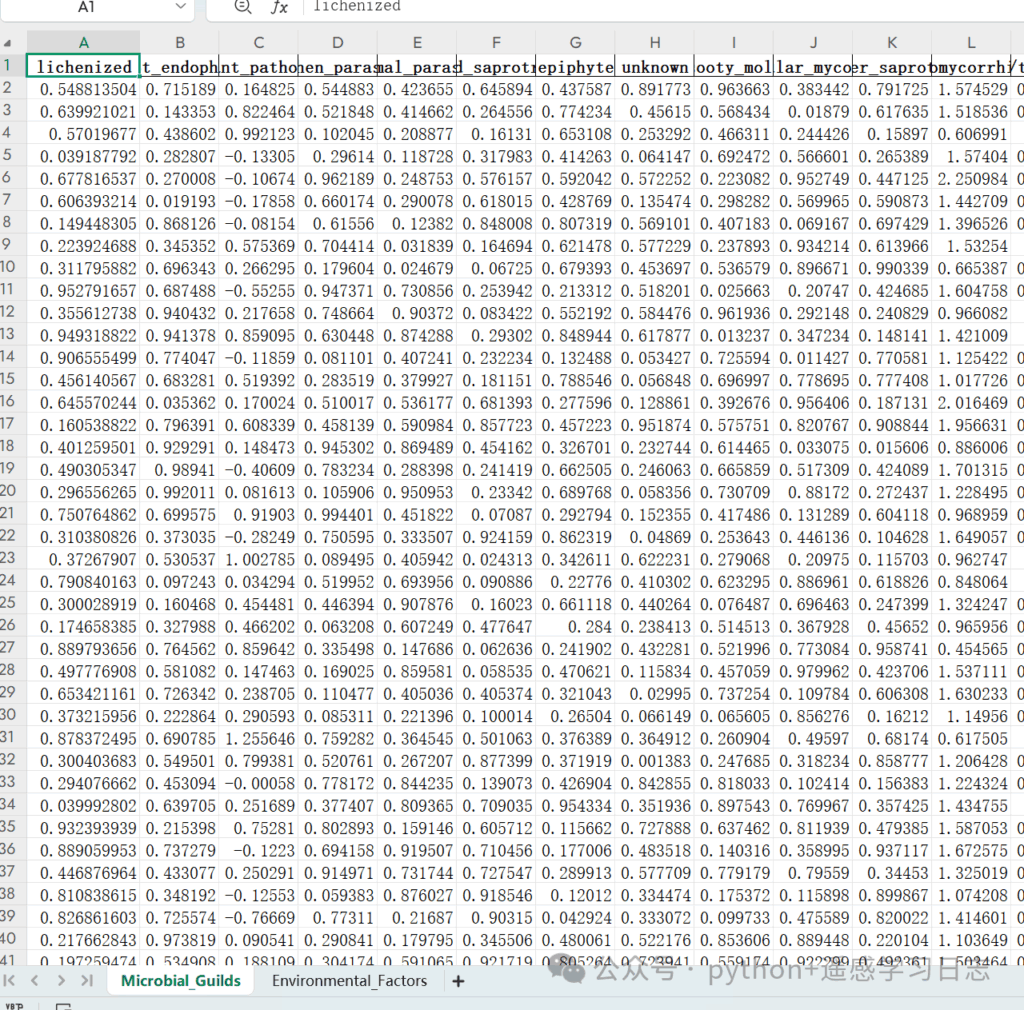
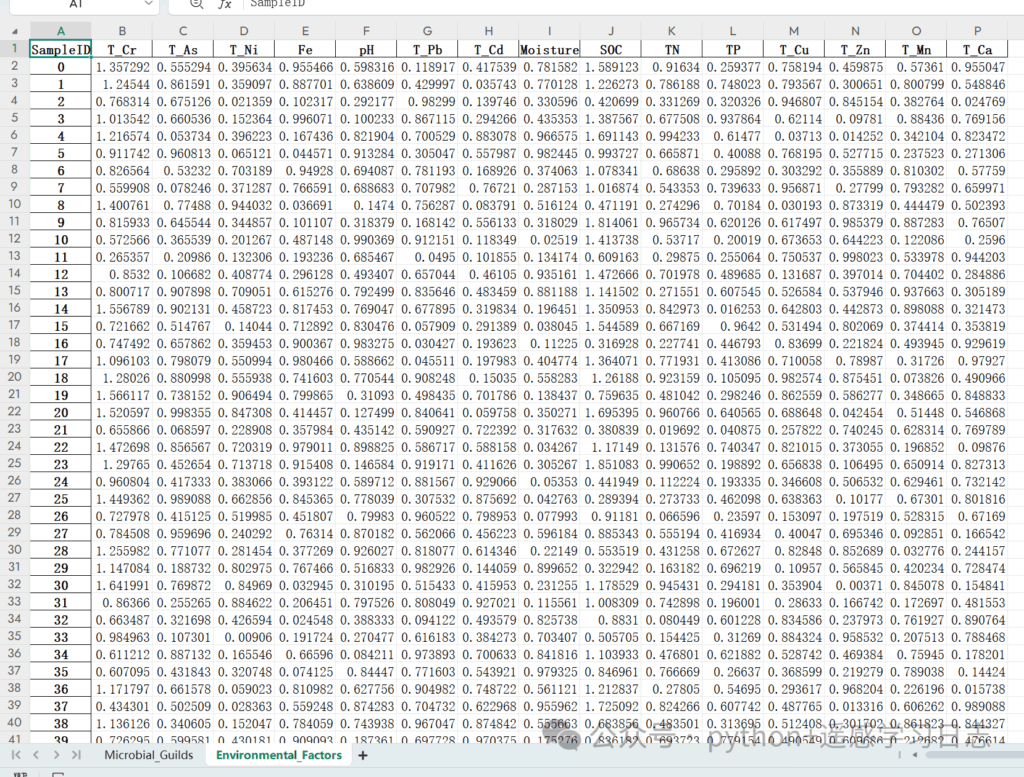
- Prepare the Excel file with two worksheets:
- Sheet 1 (e.g.,
Microbial_Guilds): rows = samples; columns = taxa/guilds; first column = SampleID - Sheet 2 (e.g.,
Environmental_Factors): rows = samples; columns = environmental variables; first column = SampleID
Ensure the SampleIDs match across sheets (same spelling & case).
- Sheet 1 (e.g.,
- Edit the main script:
input_excel_path = r'...\your_data.xlsx'sheet_name_1,sheet_name_2to your sheet names- If SampleID is not in the first column, adjust
index_colaccordingly.
- Run the script. It will:
- Align shared samples
- Compute all pairwise Spearman correlations and p-values
- Plot a clustered heatmap with significance stars
- Save a high-res PNG (
clustered_correlation.pngby default)
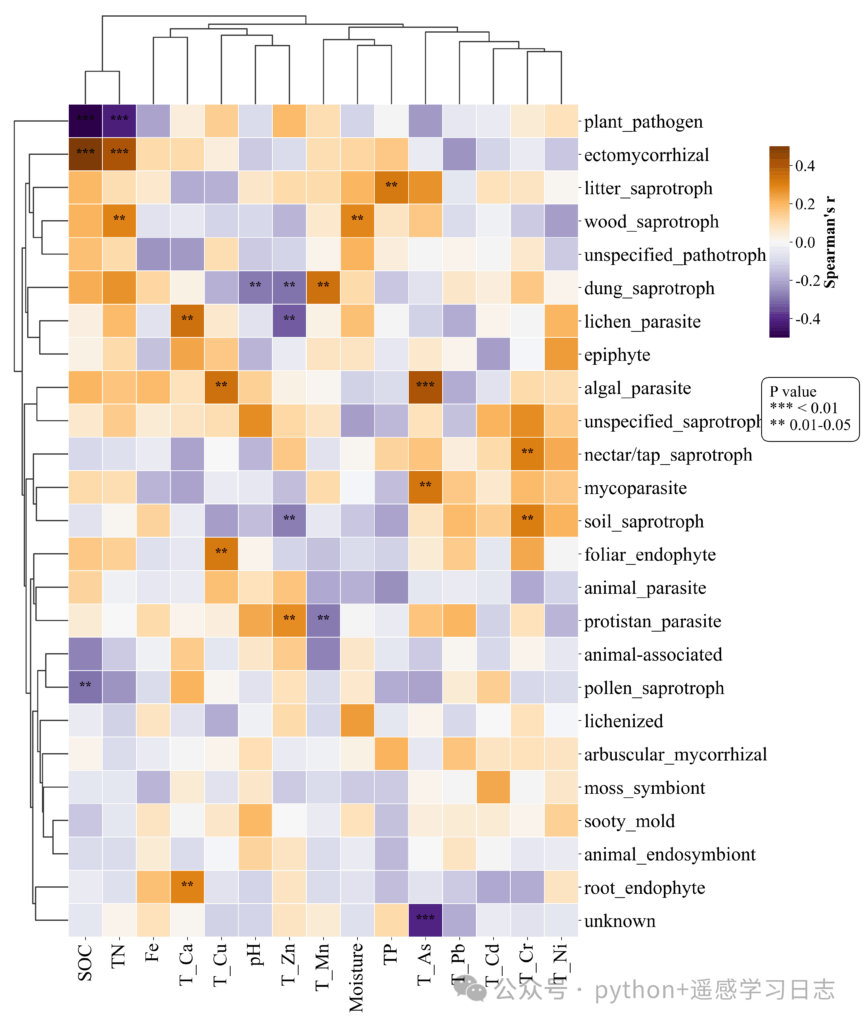
Reading the figure
- Color: correlation strength and sign (symmetric scale by default, e.g., −0.5 to +0.5).
- Stars:
***for p < 0.01,**for 0.01–0.05; blank means not significant at these thresholds. - Dendrograms: variables grouped by similarity (based on Euclidean distance over the correlation profiles). Clusters may reveal functional guilds or environmental gradients.
Practical tips
- Multiple testing: with many pairs, consider FDR control (e.g.,
statsmodels.stats.multitest.multipletests) and replace stars accordingly. - Scaling the color range: adjust
vmaxto emphasize stronger correlations (e.g.,vmax=0.7). - Reproducibility: set a random seed only if you bootstrap or sample; the clustering here is deterministic given the data.
- Performance: with thousands of variables, compute time and figure size grow quickly; consider subsetting or pre-clustering.